An inherent drawback of the traditional diffusion tensor model is its limited ability to provide detailed information about multidirectional fiber architecture within a voxel. This leads to erroneous fiber tractography results in locations where fiber bundles cross each other. This may lead to the inability to visualize clinically important tracts such as the lateral projections of the corticospinal tract. In this report, we present a deterministic two-tensor eXtended Streamline Tractography (XST) technique, which successfully traces through regions of crossing fibers. We evaluated the method on simulated and in-vivo human brain data, comparing the results with the traditional single-tensor and with a probabilistic tractography technique. By tracing the corticospinal tract and correlating with fMRI-determined motor cortex in both healthy subjects and patients with brain tumors, we demonstrate that two-tensor deterministic streamline tractography can accurately identify fiber bundles consistent with anatomy and previously not detected by conventional single tensor tractography. When compared to the dense connectivity maps generated by probabilistic tractography, the method is computationally efficient and generates discrete geometric pathways that are simple to visualize and clinically useful. Detection of crossing white matter pathways can improve neurosurgical visualization of functionally relevant white matter areas.
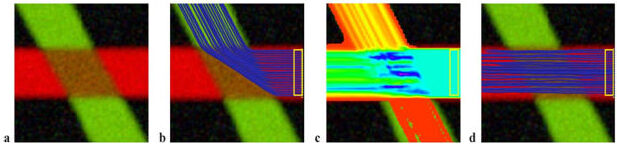
Simulated data. (a) The simulated 60° fiber crossing. Inside the brown region (middle), the two fibers are crossing each other. Outside the region each tensor is isotropic and the direction is color-coded. (b) Single tensor tractography when seeded in the region bounded by the yellow box. (c) Connection map from probabilistic tractography when seeded in the same region (voxels are color coded from 5000 (blue) to 4 (red) samples passing through the voxel). (d) Tractography based on XST.
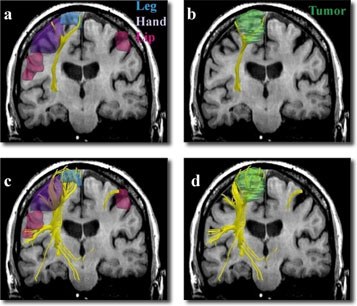
Single (a,b) vs. two-tensor XST (c,d) tractography results on a 64-year old caucasian female with left frontoparietal meningioma. Fibers were seeded in the internal capsule and those intersecting fMRI activations were retained. Three-dimensional surface models represent (a, c) fMRI areas for the foot (blue), hand (purple) and the face (pink) and (b, d) tumor (green).